Generalizable EHR-R-REDCap Pipeline for a National Multi-Institutional Rare Tumor Patient Registry
Metrics
Article and Authorship Details
Journal: JAMIA Open. Published January 7, 2022. Please see the publisher’s version on the JAMIA Open website. Copyright © 2022, Oxford University Press.
Article type: Application Note.
Authors: Sophia Z. Shalhout1 PhD, , Farees Saqlain3 , Kayla Wright1 , Oladayo Akinyemi1 , David M. Miller1,2,3* MD PhD,
1Department of Medicine, Division of Hematology/Oncology, Massachusetts General Hospital, Boston, MA
2Department of Dermatology, Massachusetts General Hospital, Boston, MA
3Harvard Medical School, Boston, MA
*Corresponding author: David M. Miller MD PhD
Massachusetts General Hospital
Email: dmiller4@mgh.harvard.edu
Short title: EHR-R-REDCap Pipeline for Patient Registries
Funding sources: The Harvard Cancer Center Merkel Cell Carcinoma patient registry is supported by Project Data Sphere. SZS was supported by the Massachusetts General Hospital Cutaneous Oncology Fellowship. Conflict of Interest Disclosure: DMM has received honoraria for participating on advisory boards for Checkpoint Therapeutics, EMD Serono, Pfizer, Merck, Regeneron and Sanofi Genzyme. The remaining authors report no competing interests.
Keywords: EHR, Rare tumor registry, Merkel Cell Carcinoma, REDCap, R statistical software, patient registries
Acknowledgements: We would like to acknowledge Ravikumar Komandur, PhD, Project Director at Project Data Sphere for assistance during manuscript preparation.
Abbreviations: BUN: Blood urea nitrogen, DD: Data Dictionary, eGFR: estimated glomerular filtration rate, EHR: Electronic Health Record, eLAB: EHR Labs Abstraction, EDW: Electronic Health Record Data Warehouse, IRB: Institutional Review Board, Labs: Laboratory values, LOINC: Logical Observation Identifiers Names and Codes, MCC: Merkel Cell Carcinoma, MCCPR: Merkel Cell Carcinoma Patient Registry, MCV: Mean corpuscular volume, MGB: Mass General Brigham, MRN: Medical record number, OS: Overall Survival, OSH: Outside Hospital, PDS: Project Data Sphere, REDCap: Research Electronic Data Capture, record_id: patient record identification number
ABSTRACT
Objective: To develop a clinical informatics pipeline designed to capture large-scale structured EHR data for a national patient registry.
Materials and Methods: The EHR-R-REDCap pipeline is implemented using R-statistical software to remap and import structured EHR data into the REDCap-based multi-institutional Merkel Cell Carcinoma (MCC) Patient Registry using an adaptable data dictionary.
Results: Clinical laboratory data were extracted from EPIC Clarity across several participating institutions. Labs were transformed, remapped and imported into the MCC registry using the EHR labs abstraction (eLAB) pipeline. Forty-nine clinical tests encompassing 482,450 results were imported into the registry for 1,109 enrolled MCC patients. Data-quality assessment revealed highly accurate, valid labs. Univariate modeling was performed for labs at baseline on overall survival (N=176) using this clinical informatics pipeline.
Conclusion: We demonstrate feasibility of the facile eLAB workflow. EHR data is successfully transformed, and bulk-loaded/imported into a REDCap-based national registry to execute real-world data analysis and interoperability.
LAY SUMMARY
Healthcare data collected during routine clinical care in patient electronic health records can be curated and organized into disease-focused registries. For example, the Merkel Cell Carcinoma Patient Registry is a multi-institutional national database that utilizes real-world health care data to inform best practices, improve patient outcomes, and test hypotheses that require large data samples. However, many challenges arise when capturing patient-level details/data from the health records especially when this data must be aggregated across multiple institutions. Here, we describe a pipeline that pulls large-scale data from the electronic health records (EHR), wrangles and remaps the data in a format appropriate for the database using R statistical software, and imports the data in bulk for thousands of patients at a time into REDCap, the electronic data capture system that houses the data. The EHR-R-REDCap pipeline can be utilized to capture many patient data features from the participating institutions. Here, we demonstrate eLAB, the EHR-R-REDCap pipeline utilized by our Merkel Cell Carcinoma Patient Registry to capture thousands of laboratory values from the EHR for the registry. We provide source code, examples, and instructions for adaptability for other clinical research projects that aim to collect large scale laboratory data from the EHR.
INTRODUCTION:
Patient registries focus on the collection of clinically relevant data around a target population. In the era of precision medicine, national patient registries have gained momentum1,2,3. Furthermore, there exists a critical need to develop rare tumor patient registries to better characterize the natural history of these cancers4,5,6. The national Merkel Cell Carcinoma (MCC) Patient Registry (MCCPR) was developed to record outcomes and events in patients diagnosed with this rare and aggressive skin cancer7,8. This will enable multiple investigators to examine real-world data to improve patient outcomes and identify best practices. The development of the MCCPR is an evolving effort led by researchers and clinical investigators from academia, industry and regulatory science, as well as patient advocates. Data is captured at participating sites using registry-specific instruments developed in the Research Electronic Data Capture (REDCap) system, a widely used web-based platform9,10. The goal is to disseminate the multi-institutional aggregated data on the first publicly held national MCCPR, utilizing Project Data Sphere’s open-access platform7.
Figure 1: Schematic overview of the multi-institutional MCCPR
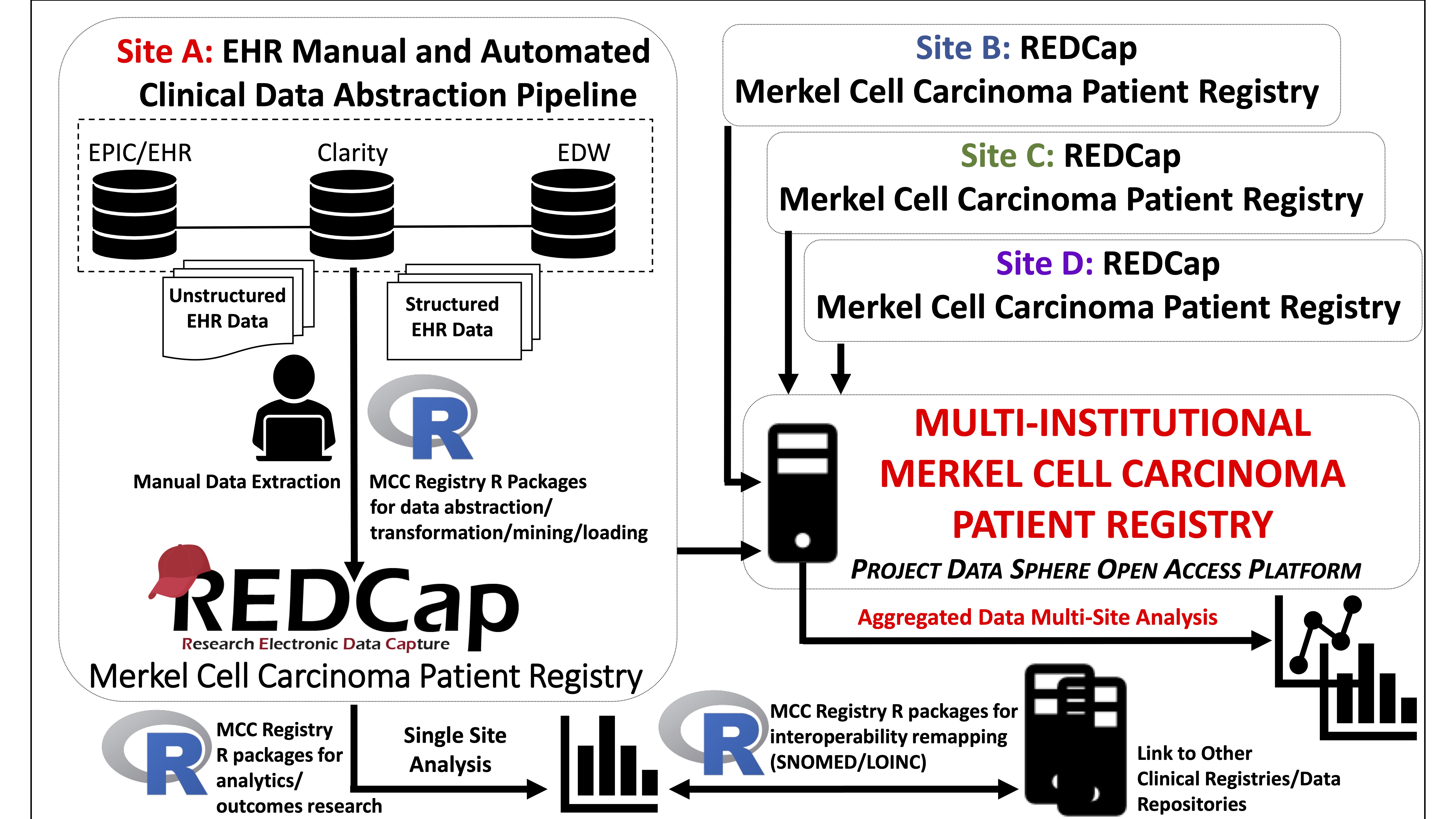
Registry data collection is a time-intensive process requiring electronic health record (EHR) navigation and manual chart review. Automated abstraction of structured EHR data allows for the rapid generation of large-scale data to drive registry research. We adopted accessible, affordable software, REDCap and R, to lower technical and economic barriers for participating sites9,10,11 to remap and load/import data into the REDCap-based MCCPR using the data dictionary (DD). The pipeline facilitates large-scale registry data collection, analysis, and interoperability/standardization for many features including demographics, and medications. Importantly, this pipeline anticipates and allows for modifications of the scripts/packages as site-specific needs arise to accommodate variability in the sources/format of initial EHR data input.
The objective of this study is to demonstrate and evaluate the EHR-R-REDCap pipeline with the EHR labs abstraction (eLAB) pipeline, developed for importing structured EHR clinical lab data using the registry REDCap DD. We implemented eLAB to extract lab data from the EHR of 1,109 registry subjects from several institutions including Massachusetts General Hospital, Dana-Farber Cancer Center, Brigham and Women’s Hospital and several satellite community sites. We also performed proof-of-concept exploratory outcomes research on a test cohort (N =176). Finally, we evaluated the capture time and data quality/accuracy across patients from different sites in comparison to manually abstracted data and the EHR. We developed and provide here: (1) the data dictionary (DD) to allow users to adopt the MCCPR REDCap labs instrument, (2) key-value/lookup tables for remapping ~300 EHR lab-subtypes into the 35 registry-specific labs of interest (3) eLAB source code, and (4) a sample data set for demonstration purposes. Furthermore, we provide (5) a detailed dynamic report as an R-markdown12 file annotating all functions/R-script. We highlight examples of upfront script modifications that may be needed to accommodate bulk lab data pulls from other EHR data warehouse (EDW) sites/sources. Downstream script is largely preserved when utilizing the accompanying. These resources may be adopted by other clinical research projects/registries to minimize developer efforts. We outline a pipeline for multi-institutional sites to aggregate large-scale data under institutional review board (IRB)-approved studies.
MATERIALS AND METHODS
eLAB Development and Source Code (R statistical software):
eLAB is written in R11 (version 4.0.3), and utilizes the following packages for processing: DescTools13, REDCapR14, reshape215, splitstackshape16, readxl17, survival18, survminer19, and tidyverse20. Source code for eLAB can be downloaded directly. eLAB reformats EHR data abstracted for an identified population of patients (e.g. medical record numbers (MRN)/name list) under an Institutional Review Board (IRB)-approved protocol. The MCCPR does not host MRNs/names and eLAB converts these to MCCPR assigned record identification numbers (record_id) before import for de-identification. Functions were written to remap EHR bulk lab data pulls/queries from several sources including Clarity/Crystal reports or institutional EDW including Research Patient Data Registry (RPDR) at Mass General Brigham (MGB). The input, a csv/delimited file of labs for user-defined patients, may vary. Thus, users may need to adapt the initial data wrangling script based on the data input format. However, the downstream transformation, code-lab lookup tables, outcomes analysis, and LOINC remapping are standard for use with the REDCap DD. The available R-markdown provides suggestions and instructions on where or when upfront script modifications may be necessary to accommodate input variability.
The eLAB pipeline takes several inputs. For example, the input for use with the ‘ehr_format(dt)’ single-line command is non-tabular data assigned as R object ‘dt’ with 4 columns: 1) Patient Name (MRN), 2) Collection Date, 3) Collection Time, and 4) Lab Results wherein several lab panels are in one data frame cell. A mock dataset in this ‘untidy-format’ is provided for demonstration purposes. (Figure 2, Supplemental Materials).
Figure 2: Schematic Overview of the eLAB Clinical Informatics Pipeline.
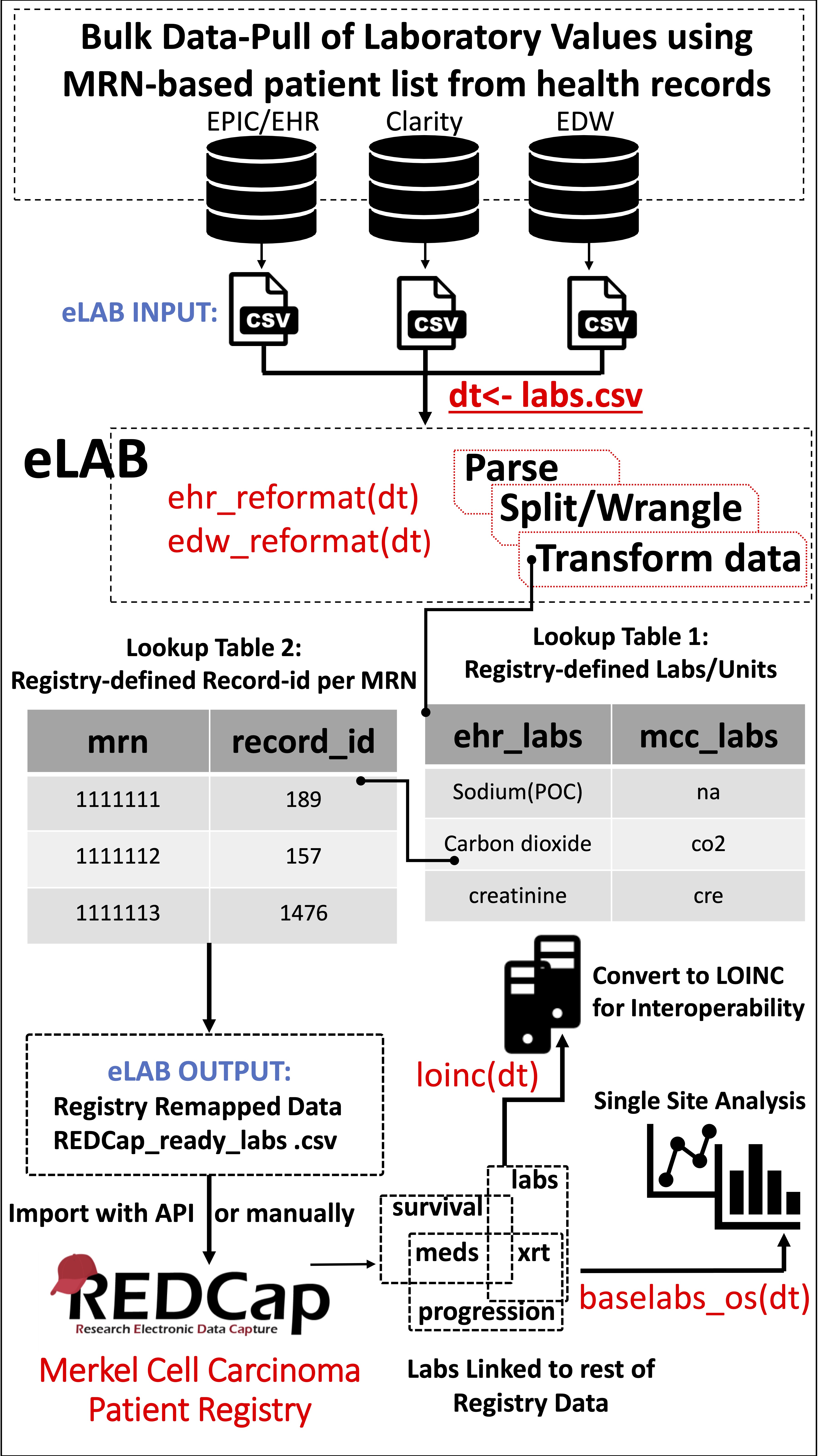 Figure 2: Schematic Overview of the eLAB Clinical Informatics Pipeline. eLAB is designed to take as input csv data from several EHR/EDW sources and may be adapted for other site-specific inputs. Once delimited data is assigned as R object ‘dt’, a single-line command is used to transform the data into the REDCap-ready registry configuration for import based on source type (e.g. ehr_reformat(), or edw_reformat()). eLAB is designed to de-identify the patient names/medical record numbers (MRN) with registry specific record identification (record_id) numbers. Furthermore, 300 subtypes of laboratory tests (ehr_labs) are remapped into 35 registry fields (mcc_labs). Once imported into the registry via an output csv file or using REDCap API token, single-line commands are used for outcomes research and analysis (baselabs_os()), as well as remapping for interoperability with standardized SNOMED or LOINC code (loinc()). LOINC remapping is an optional feature provided by eLAB to allow the MCCPR data to be linked to other non-MCCPR clinical research efforts/ registries that may utilize LOINC. Using a look-up table dependent on the MCCPR data dictionary, eLAB remaps 1 LOINC code per 1 Data dictionary field variable name only after the data has successfully been cleaned, remapped, transformed and imported.
Figure 2: Schematic Overview of the eLAB Clinical Informatics Pipeline. eLAB is designed to take as input csv data from several EHR/EDW sources and may be adapted for other site-specific inputs. Once delimited data is assigned as R object ‘dt’, a single-line command is used to transform the data into the REDCap-ready registry configuration for import based on source type (e.g. ehr_reformat(), or edw_reformat()). eLAB is designed to de-identify the patient names/medical record numbers (MRN) with registry specific record identification (record_id) numbers. Furthermore, 300 subtypes of laboratory tests (ehr_labs) are remapped into 35 registry fields (mcc_labs). Once imported into the registry via an output csv file or using REDCap API token, single-line commands are used for outcomes research and analysis (baselabs_os()), as well as remapping for interoperability with standardized SNOMED or LOINC code (loinc()). LOINC remapping is an optional feature provided by eLAB to allow the MCCPR data to be linked to other non-MCCPR clinical research efforts/ registries that may utilize LOINC. Using a look-up table dependent on the MCCPR data dictionary, eLAB remaps 1 LOINC code per 1 Data dictionary field variable name only after the data has successfully been cleaned, remapped, transformed and imported.
Bulk lab data pulls often result in subtypes of the same lab. For example, potassium labs are reported as “Potassium,” “Potassium-External,” “Potassium(POC),” “Potassium,whole-bld,” “Potassium-Level-External,” “Potassium,venous,” and “Potassium-whole-bld/plasma.” eLAB utilizes a key-value lookup table with ~300 lab subtypes for remapping labs to the DD code. eLAB reformats/accepts only those lab units pre-defined by the registry DD. The lab lookup table is provided for direct use or may be re-configured/updated to meet end-user specifications. eLAB is designed to remap, transform, and filter/adjust value units of semi-structured/structured bulk laboratory values data pulls from the EHR to align with the pre-defined code of the DD.
Data Dictionary (DD)
EHR clinical laboratory data is captured in REDCap using the ‘Labs’ repeating instrument (Supplemental Figure 2). The DD is provided for use by researchers at REDCap-participating institutions and is optimized to accommodate the same lab-type captured more than once on the same day for the same patient. The instrument captures 35 clinical lab types (Table 1). The DD serves several major purposes in the eLAB pipeline. First, it defines every lab type of interest and associated lab unit of interest with a set field/variable name. It also restricts/defines the type of data allowed for entry for each data field, such as a string or numerics. The DD is uploaded into REDCap by every participating site/collaborator and ensures each site collects and codes the data the same way. Automation pipelines, such as eLAB, are designed to remap/clean and reformat data/units utilizing key-value look-up tables that filter and select only the labs/units of interest. eLAB ensures the data pulled from the EHR contains the correct unit and format pre-configured by the DD. The use of the same DD at every participating site ensures that the data field code, format, and relationships in the database are uniform across each site to allow for the simple aggregation of the multi-site data. For example, since every site in the MCCPR uses the same DD, aggregation is efficient and different site csv files are simply combined (Figure 3).
Figure 3: eLAB and the Data Dictionary to Harmonize Multi-Institutional Data Aggregation.
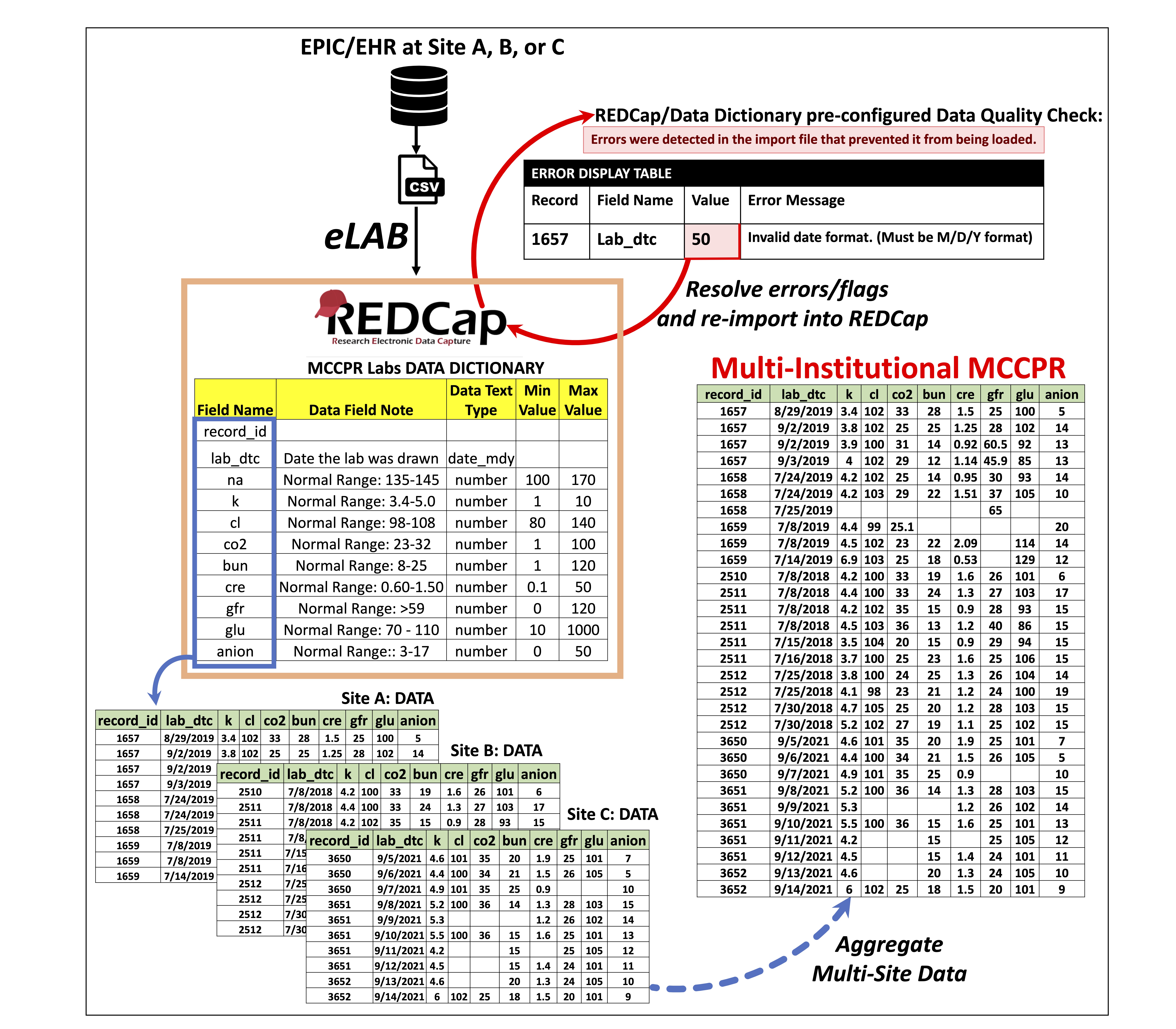 Figure 3: eLAB and the Data Dictionary to Harmonize Multi-Institutional Data Aggregation. The data dictionary, once uploaded into REDCap, creates the lab capture system or the “Labs instrument.” eLAB is designed to reformat and normalize laboratory values and units that are bulk- pulled from the EHR. eLAB transforms the data into a format pre-defined by the data dictionary and its associated variable codes. eLAB performs the primary data cleansing steps. However, one final quality check is also utilized at the very end during import of data. If an attempt is made to import any data that is incorrectly reformatted (ie. numerical value for a date field that is not in the acceptable M/D/Y format), an error is set off during the final stage of import by the REDCap scaffold that is associated with the pre-configured design of the data dictionary. Data with errors will fail to import into REDCap. The REDCap data import tool will display the data that does not conform to the configuration designated by the data dictionary and it will be flagged, alongside error messages that provide guidance on how to resolve the issue. The data will have to be re-evaluated and corrected. Only when the data is free of errors and all issues are resolved, can a successful upload/import into REDCap occur. With every site in the multi-institutional registry utilizing eLAB and the exact same data dictionary, aggregation of the data is straightforward and only requires combining/appending the outputs of each site together (ie each site will combine individual ‘.csv file’ into one large multi-site ‘.csv file’ for the final aggregated data.) Multi-site data aggregation is facilitated by each participating site 1) utilizing eLAB for transformation and normalization with 2) the accompanying data dictionary.
Figure 3: eLAB and the Data Dictionary to Harmonize Multi-Institutional Data Aggregation. The data dictionary, once uploaded into REDCap, creates the lab capture system or the “Labs instrument.” eLAB is designed to reformat and normalize laboratory values and units that are bulk- pulled from the EHR. eLAB transforms the data into a format pre-defined by the data dictionary and its associated variable codes. eLAB performs the primary data cleansing steps. However, one final quality check is also utilized at the very end during import of data. If an attempt is made to import any data that is incorrectly reformatted (ie. numerical value for a date field that is not in the acceptable M/D/Y format), an error is set off during the final stage of import by the REDCap scaffold that is associated with the pre-configured design of the data dictionary. Data with errors will fail to import into REDCap. The REDCap data import tool will display the data that does not conform to the configuration designated by the data dictionary and it will be flagged, alongside error messages that provide guidance on how to resolve the issue. The data will have to be re-evaluated and corrected. Only when the data is free of errors and all issues are resolved, can a successful upload/import into REDCap occur. With every site in the multi-institutional registry utilizing eLAB and the exact same data dictionary, aggregation of the data is straightforward and only requires combining/appending the outputs of each site together (ie each site will combine individual ‘.csv file’ into one large multi-site ‘.csv file’ for the final aggregated data.) Multi-site data aggregation is facilitated by each participating site 1) utilizing eLAB for transformation and normalization with 2) the accompanying data dictionary.
Study Cohort
This study was approved by the MGB IRB Protocol# 2019P002459. Search of the EHR was performed to identify patients diagnosed with MCC between 1975-2021 (N=1,109) for inclusion in the MCCPR. Subjects diagnosed with primary cutaneous MCC between 2016-2019 (N= 176) were included in the test cohort for exploratory studies of lab result associations with overall survival (OS) using eLAB.
Statistical Analysis
OS is defined as the time from date of MCC diagnosis to date of death. Data was censored at the date of the last follow-up visit if no death event occurred. Univariable Cox proportional hazard modeling was performed among all lab predictors. Due to the hypothesis-generating nature of the work, p-values were exploratory and Bonferroni corrections were not applied.
RESULTS:
Clinical labs for 1109 registry subjects were extracted from EPIC Clarity and produced >500,000 lab values. eLAB was utilized to remap results using the REDCap DD, de-identify the patients with a pre-defined MCCPR ‘record_id’, and remove non-importable characters such as ‘refused,’ and ‘canceled.’ Lab subtypes were remapped under the MCCPR lab categories and transformed into a REDCap-ready importable configuration. 482,450 lab values for 1109 patients were successfully imported.
We assessed the accuracy of the eLAB pipeline by evaluating the imported results compared to those manually collected by two registry-trained data abstractors. REDCap start/stop date-timestamp fields were implemented to measure time directly spent on manually abstracting structured lab data, including EHR navigation time. Abstractors spent a total of 1,458.4 minutes collecting 8,043 lab values on 30 patients across the different sites. On average, 5.5 laboratory values per minute were abstracted from EHR. In comparison, ~3x as many values for the same 30 patients were transformed, remapped and imported using eLAB in 1.2 minutes. A 97.6% agreement rate was achieved when the two datasets were analyzed for non-agreement, with manual number/typo/date errors accounting for the differences as manually confirmed in the EHR to determine the dataset with the error.
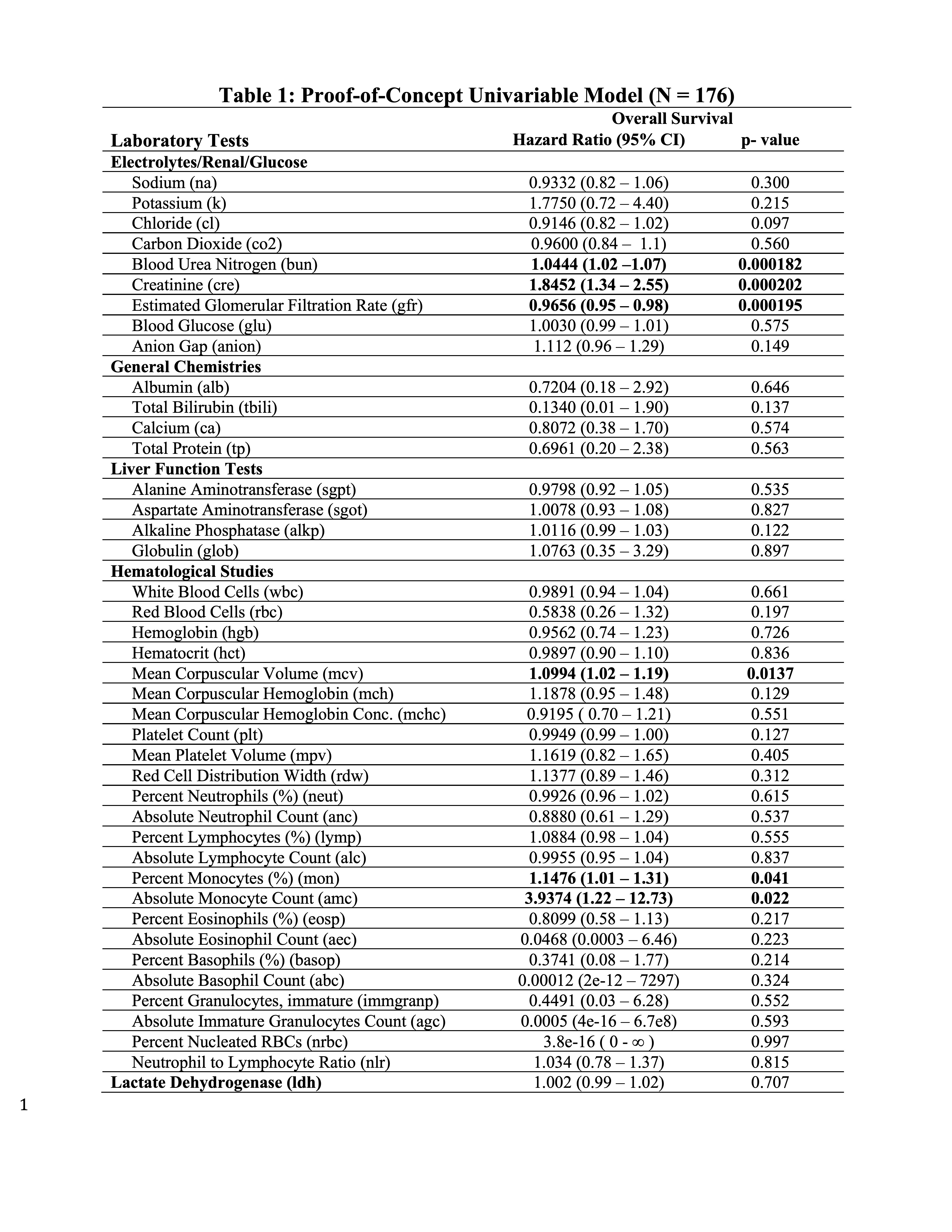
Large-scale registry data aggregation is useful for the identification of predictive/prognostic biomarkers, and to uncover patient disease susceptibility. Therefore, proof-of-concept exploratory univariate modeling was performed with eLAB for each lab with date of diagnosis set to baseline for a single-site test cohort (N =176) (Table 1). eLAB calls the appropriate registry survival data from the ‘subject-status’ instrument, date of diagnosis in the ‘patient characteristics instrument’ and date cutoffs for laboratory data to perform Cox proportional Hazards modeling. BUN, creatinine, eGFR, MCV, as well as percent/absolute monocyte count were found associated with OS in patients with MCC in the test cohort (p- values < 0.05, Table 1).
Table 1: Proof-of-Concept Univariable Model.
DISCUSSION:
Multi-institutional patient registries are critical in optimizing clinical management and reducing the bias inherent in single-institutional studies. A major barrier for developing national and international programs is the rapid, consistent collection of large-scale data allowing data aggregation and analysis. Here, we develop and evaluate the MCCPR EHR-R-REDCap pipeline using eLAB across several participating sites. In addition, the successful remapping, transformation, import and analysis of clinical laboratory data on 176 patients was carried out in a fraction of the time required by the standard manual entry pipeline. The time required for manual data capture for 30 patients alone was >1000x fold longer. There are caveats to this time-to-capture comparison. For example, while manual abstraction compared directly to our final automated product is much slower, code development and refinement is a lengthy process and not accounted for in the time-to-capture comparison. Our registry key-value/lookup tables are rarely updated, but if they are often redesigned and re-curated during one’s clinical research efforts, it may increase automated time-to-capture. Furthermore, much of the data capture time required by the EHR-R-REDCap pipeline is dedicated to the importation of the data into REDCap and depends heavily on internet latency. Finally, this system is designed to augment manual abstraction of EHR laboratory data. For example, manual abstractors may instead, focus efforts on capturing unstructured lab data from OSH faxed/scanned reports.
Several strategies have been developed and proposed in the past to automate the retrieval of structured EHR data for import into REDCap repositories including dynamic data pull modules21,22, and REDCap plugins that connect REDCap directly to the EHR and SFTPs servers15. However, these strategies often require dedicated institutional REDCap personnel to build web services as middleware and require data format setup. Selections are often negotiated with the institution’s REDCap administrators. In addition, each participating site will need to individually remap sourced data to the registry fields. In the EHR-R-REDCap pipeline, the registry defined acceptable source values are provided for participating sites, and users may adjust/add as needed. With eLAB, several EHR values are easily mapped to the same registry code when appropriate and has proven successful across several sites.
Furthermore, proposed methods that rely on REDCap-EHR direct communication fetch data in real-time and need to be adjudicated per value per patient, before importing the data into the project. The EHR-R-REDCap pipeline takes advantage of REDCap’s data import scaffold, allowing for the identification of errors in bulk and user-adjudication before overwriting any previous data stored in the registry. The EHR-R-REDCap pipeline allows for routine bulk pulls during user-defined time windows. For example, in longitudinal studies where data points are routinely updated such as laboratory values, data can be systematically updated once a month only, on all patients simultaneously in the registry. Finally, it may be challenging for each participating institution, including community sites, to map individual institution’s data models to standardized codes and with rare tumors, each site is essential to increase enrollment.
The EHR-R-REDCap pipeline also reduces the barriers for data wrangling and analysis at each site. We demonstrate feasibility of eLAB to carry out exploratory data analysis (Table 1). Conclusions related to MCC are beyond the scope of this work but will be addressed in additional single- and multi- institutional studies.
CONCLUSIONS:
In summary, the MCCPR EHR-R-REDCap pipeline may help other multi-institutional registries/studies collect, aggregate, and analyze data rapidly. We provide the REDCap instrument DD, eLAB source code with instructions, key-value lookup tables as well as a simulated data set to the research community. Notably, an annotated R-markdown detailing where end-users may need to adjust upfront script is also provided to assist with modifications/adaptability at other sites depending on the input format of the data source.
SUPPLEMENTAL METHODS
Supplemental Methods and Figures.
EHR Labs Abstraction Pipeline (eLAB) Details: eLAB was developed in R (version 4.0.3) to re-configure structured data extracted from the EHR for REDCap-ready import into registries/repositories utilizing the provided pre-configured DD. eLAB takes as input bulk lab data pulled from the EHR. The lab data is then parsed, split, wrangled, filtered, and transformed by eLAB script on only those specific labs that are collected by our registry based on the key-value match/filtering table. eLAB remaps the labs data and units using the key-value match/filtering table and retains the labs of interest and the units of interest accepted by our registry data dictionary. Furthermore, eLAB only keeps strings/formats acceptable by our registry data dictionary. For example, script in eLAB is designed to exclude those labs with certain results such as “REFUSED LAB”/”CANCELLED” when a numeric value is pre-designated as the appropriate data type for that lab type.
The provided source code is fully annotated to describe how each of these steps is performed by the relevant eLAB script. First, eLAB requires the bulk-pulled lab data to be loaded into the R environment and then renames the csv file as the object, “dt” in R (Figure 2). An accompanying sample “bulk EHR” dataset for user testing is also provided. After loading and renaming the data, eLAB script reconfigures columns with dates and times in the data table as ‘Y/M/D/H/M/S’ format. eLAB then splits and parses the data by separating units from values based on delimiters. Data is then remapped and filtered based on look-up tables to retain only labs and units the registry collects. These are pre-defined by the accompanying DD and eLAB reconfigures outputs/units not accepted by the registry. The data is then de-identified by using a key-value lookup table to remove patient names/medical record numbers with record identification numbers (Figure 2). Sample de-identification look-up tables are also provided for the end user to test eLAB. Final cleaning of data and data pivot from long-to-wide format transforms the data to match the strict REDCap-ready importation rules/allowances. If no errors are detected, data can be successfully imported into REDCap. Users are provided the DD for setup at their institutional REDCap site. Once imported, eLAB allows analysis of lab data with survival outcomes when linked with the full registry data elements. eLAB, for example, performs univariate analysis using the R packages survival and survminer to perform cox proportional hazard modeling on labs set to baseline. Baseline is defined as labs within a month of the date of diagnosis of MCC, and the overall survival of the patient, defined as the time from date of diagnosis and death/last follow-up visit. The provided DD also contains data fields to capture these elements manually by users. By standardizing the DD, REDCap instrument, and providing the source code, this pipeline promotes interoperability for aggregated multi-institutional collaborative studies and analysis (Figure 2).
Data Dictionary (DD):
The DD is provided for use by researchers at REDCap-participating institutions and is optimized to accommodate the same lab-type captured more than once on the same day for the same patient. To aid manual data capture of unstructured data, the instrument was designed with conditional branching/skip-logic, where the lab tests not performed on a patient are hidden. Maximum/minimum limits were set to warn of entry errors. Once uploaded into the participating REDCap institution’s site, the DD creates the Labs electronic data capture system designed to house the data that eLAB reformats and transforms. Users may also use the Labs instrument for manual entry of lab value data. Furthermore, uploading the DD also allows users to manually capture the data fields required for outcomes measurements. These fields are located in the patient characteristics and subject status and include items such as date of death and date of diagnosis. (Supplemental Figure 1 and 2)
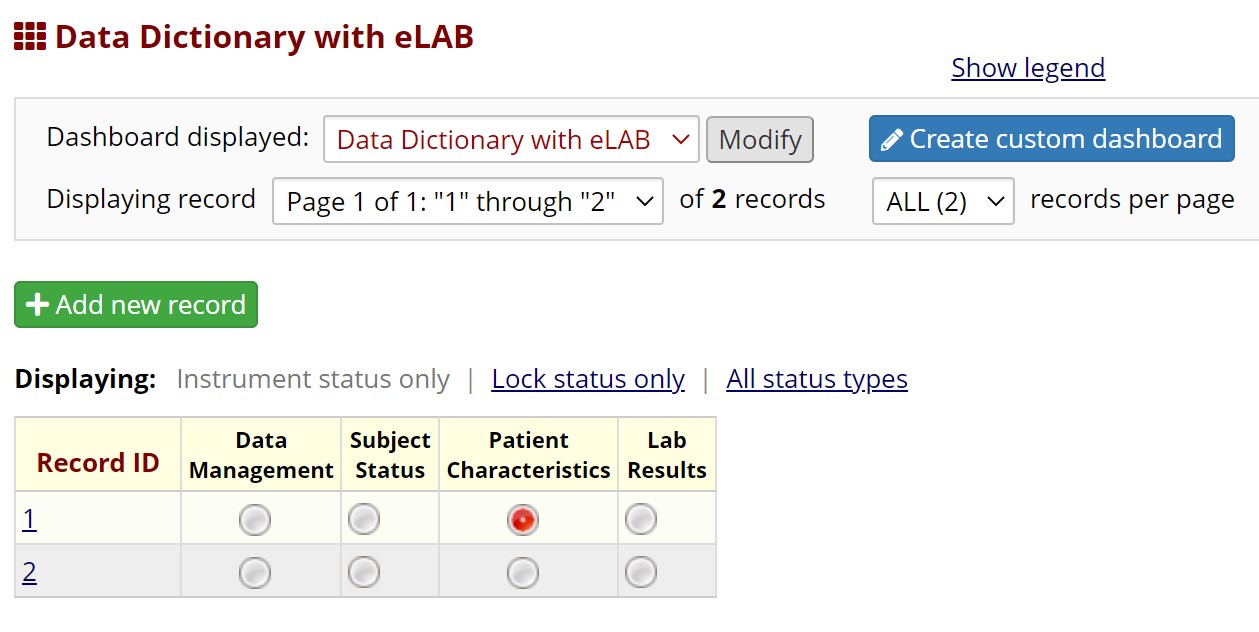 Supplemental Figure Legend 1: Users may download the eLAB data dictionary from https://github.com/TheMillerLab/eLAB (DataDictionary_eLAB.csv). The csv file should be uploaded into a REDCap project using the “Upload your Data Dictionary file (CSV file format only)” feature, at any REDCap participating institution. Choose the delimiter ‘comma’ for upload. Upon uploading, the user has to commit the changes. Shown here is the electronic data capture system in REDCap upon uploading eLAB DD. The ‘Lab Results’ instrument can be used to upload eLAB reformatted and transformed bulk data. Manual entry of lab data is also possible, and the user interface/user experience has been optimized for manual entry as well. Data fields required for outcomes measurements, including patient date of diagnosis and date of death are found in the subject status and patient characteristics instruments.
Supplemental Figure Legend 1: Users may download the eLAB data dictionary from https://github.com/TheMillerLab/eLAB (DataDictionary_eLAB.csv). The csv file should be uploaded into a REDCap project using the “Upload your Data Dictionary file (CSV file format only)” feature, at any REDCap participating institution. Choose the delimiter ‘comma’ for upload. Upon uploading, the user has to commit the changes. Shown here is the electronic data capture system in REDCap upon uploading eLAB DD. The ‘Lab Results’ instrument can be used to upload eLAB reformatted and transformed bulk data. Manual entry of lab data is also possible, and the user interface/user experience has been optimized for manual entry as well. Data fields required for outcomes measurements, including patient date of diagnosis and date of death are found in the subject status and patient characteristics instruments.
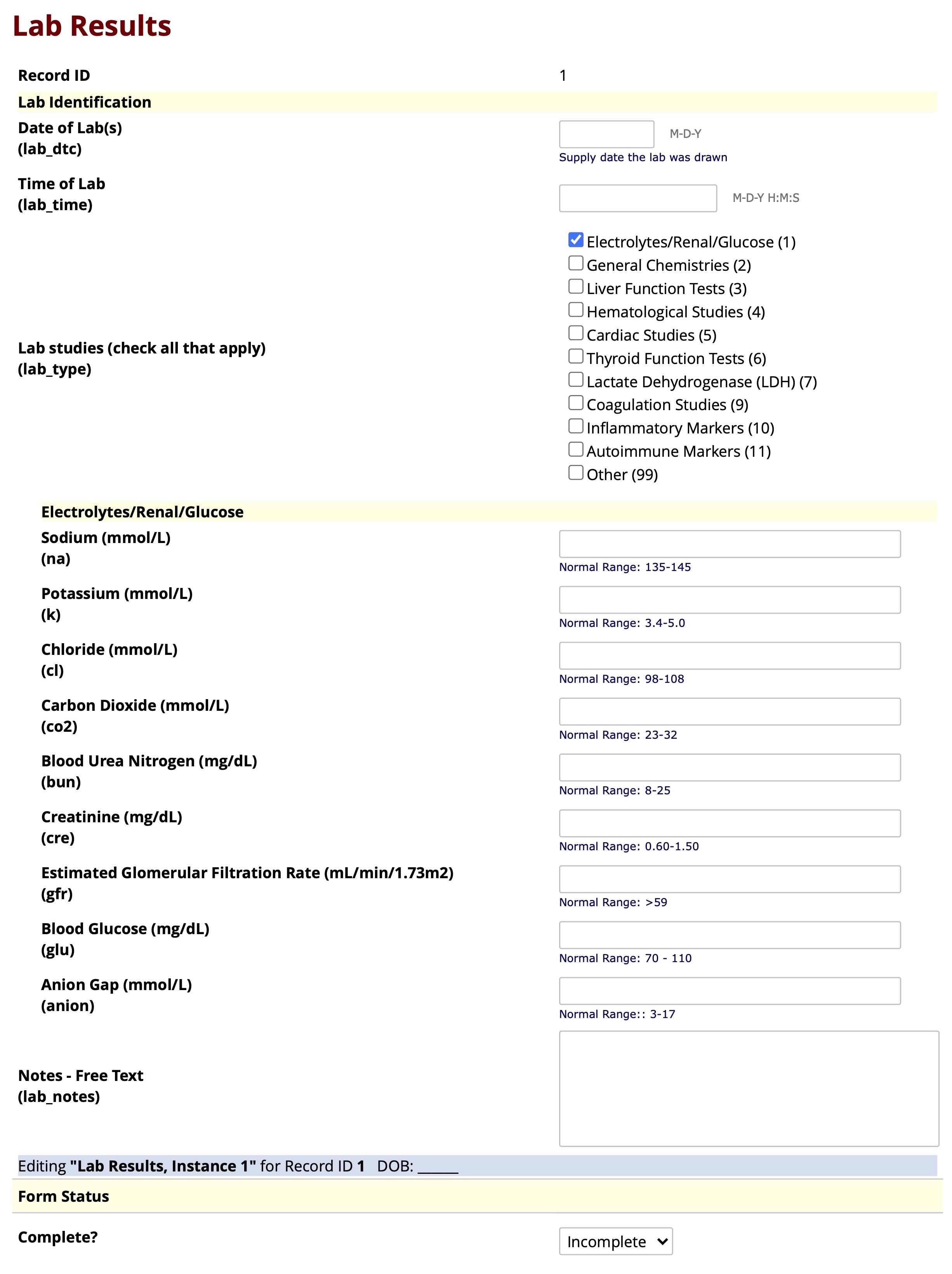 Supplemental Figure Legend 2: Shown is the user interface of the REDCap “Lab Results” instrument after uploading the eLAB data dictionary. Bulk data pulls of laboratory values from the EHR can be remapped and transformed using eLAB and then imported and housed in the REDCap ‘Lab Results’ instrument. This instrument may also be used to capture lab data manually. To aid manual data capture of unstructured data, the instrument was designed with conditional branching/skip-logic, where the lab tests not performed on a patient are hidden. For example, in this figure, only “Electrolytes/Renal/Glucose” are checked off as the current labs of interest. Therefore, branching logic only shows those relevant labs for data entry and “general chemistries” and other unchecked lab types are hidden from view.
Supplemental Figure Legend 2: Shown is the user interface of the REDCap “Lab Results” instrument after uploading the eLAB data dictionary. Bulk data pulls of laboratory values from the EHR can be remapped and transformed using eLAB and then imported and housed in the REDCap ‘Lab Results’ instrument. This instrument may also be used to capture lab data manually. To aid manual data capture of unstructured data, the instrument was designed with conditional branching/skip-logic, where the lab tests not performed on a patient are hidden. For example, in this figure, only “Electrolytes/Renal/Glucose” are checked off as the current labs of interest. Therefore, branching logic only shows those relevant labs for data entry and “general chemistries” and other unchecked lab types are hidden from view.
eLAB and Quality Assurance (QA): eLAB was created and refined over many iterations until no errors were detected during manual quality control of hundreds of data values as well as with REDCaps’s import scaffold. Quality control included manual EHR/REDCap validation of over two hundred data points across all 35 lab types across all sites, as well as double abstraction of partial charts of ~100 patients and double abstraction of full charts of 30 patients. (See https://www.themillerlab.io/post/optimizing_rwd_collection-in_instrument_qc_in_redcap/ , https://www.themillerlab.io/post/optimizing_rwd_collection-qi-initiative-error_injection_tests/ for more details about the QA methods of the MCCPR). The eLAB code was finalized for registry use when no errors were observed during validation, double abstraction, and the import flag system.